Introduction
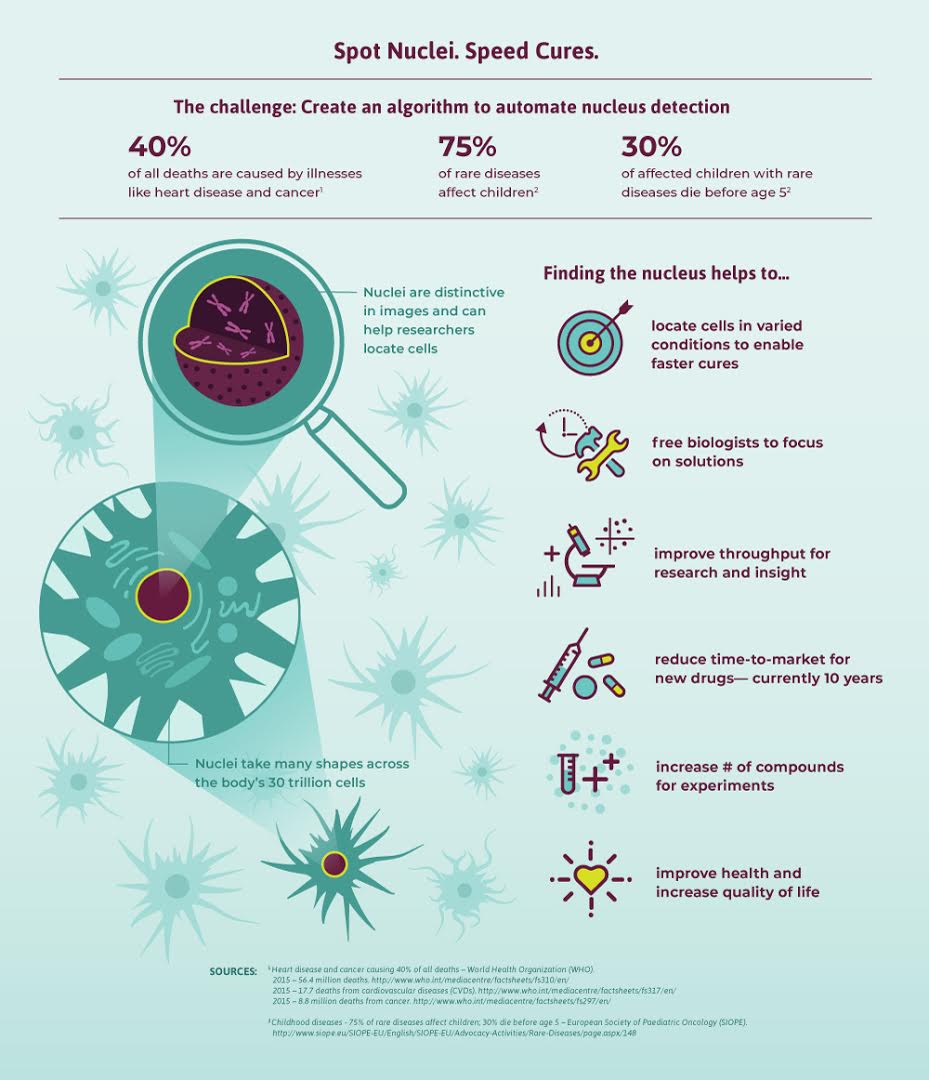
Imagine speeding up research for almost every disease, from lung cancer and heart disease to rare disorders. The first step in analyzing and curing any disease relies upon being able to first accurately detect where it is infecting, and which tissues are underlying pathological changes.
Our project is focused on segmenting one of the most important components of human cells, its nucleus. Specifically, identifying the cells’ nuclei is the starting point for most analyses because most of the human body’s 30 trillion cells contain a nucleus full of DNA, the genetic code that programs each cell. Identifying nuclei allows researchers to identify each individual cell in a sample, and by measuring how cells react to various treatments, the researcher can understand the underlying biological processes at work. Being able to do so quickly in a parallelized manner will allow clinical testing time to significantly shrink.
Figure 1: image to be segmented

Figure 2: intended segmented result
Source: 2018 Data Science Bowl challenge
Download
(attached Data Reproduction Documentation)
- 670 Training set: different cell types and tissues
- ~70 Test set: without ground truth masks
"We’ve all seen people suffer from diseases like cancer, heart disease, chronic obstructive pulmonary disease, Alzheimer’s, and diabetes. Many have seen their loved ones pass away. Think how many lives would be transformed if cures came faster. By automating nucleus detection, you could help unlock cures faster—from rare disorders to the common cold. "
Application: Big Compute + Big Data
An offline, compute intensive toolbox for nuclei segmentation
- allows fast nuclei segmentation for new images
- provides three segmentation models:
Otsu's Method
A straightforward yet effective algorithm that finds a single intensity threshold to separate all pixels into two classes, foreground and background. It completely fits our need for segmenting cell nucleus as this is exactly a two-class image segmentation problem: separating cell nucleus, and non cell nucleus, which usually differ reasonably in terms of pixel intensity (although with considerable noise in some cases).
Boruvka's Method
A well-known parallel greedy algorithm for finding a minimum spanning tree (MST) in a graph.
The algorithm employed in image segmentation treats every pixels in the image as a vertex, and edge weights measured by the dissimilarity between vertices (neighboring pixels).
Each vertex is initially placed in its own component, and the method merges regions by a criterion that the resulting segmentation is neither too coarse nor too fine.
UNet (deep learning)
A popular deep neural network that has shown great performance in image segmentation. It is a U-shaped stack of convolutional layers that up-samples a large number of local features and then concatenates with features maps in the reverse order while downsampling to the image space. Configured with a large number of trainable parameters, it is able to learn from annotated data through epochs of training and accurately determine whether each pixel belongs to a nucleus or not. With data augmentation techniques, this model learns well through only very few epochs.
Otsu's Method
Refer to the below graph for the major steps of the algorithm.

At step 0, a smiley face image is given as input.
At step 1, the algorithm goes over all pixels, and prepares a pixel intensity histogram (stored in an array).
At step 2, based on such histogram statistics, the algorithm do an exhaustive search of the best threshold that minimizes the intra-class variance, defined as a weighted sum of variances of the two classes: \({\sigma_{w}}^{2}(t) = w_0(t){\sigma_{0}}^2(t) + w_1(t){\sigma_{1}}^2(t) \). Weights \(w_1(t) = \sum_{i=0}^{t-1} p(i)\) and \(w_0(t) = \sum_{t}^{L} p(i)\) are the probabilities of the two classes separated by the threshold value of the tth bin in the histogram out of a total of \(L\) bins.
Otsu Tech stack
- - Google Cloud C2-standard-30 instance
- - Language: C++ implemented from scratch
- - Job Manager: Slurm
- - Parallel Execution Model: Single Program-Multiple Data (SPMD)
- - Type of Application: HPC
- - MPI/OpenMP Hybrids systems
- - Inter-node: MPI (distributed memory model), Data Parallelism
- - Intra-node: OpenMP (shared memory model), Data Parallelism
Boruvka’s Method
Image segmentation strives to partition a digital image into regions of pixels with similar properties. If we model an image as a graph, a simple and comparatively space-efficient variant of this is a grid graph, whereby each pixel is connected to its neighbors in the four, or eight cardinal directions. The graph is undirected because of symmetry. Then we can think about edge weight as the dissimilarity between two neighboring vertices.
Borůvka's algorithm is a greedy algorithm for finding a minimum spanning tree in a graph, or a minimum spanning forest in the case of a graph that is not connected.
The algorithm begins by in the beginning treating every vertex as a one-vertex tree, for each tree, finding the minimum-weight edge from this tree to all the other vertices of the graph, and adding all of those edges to the forest (set of all trees). Then, it repeats the process of finding the minimum-weight edge from each tree constructed so far to a different tree, and adding all of those edges to the forest. Each repetition of this process reduces the number of trees, within each connected component of the graph, to at most half of this former value, so after logarithmically many repetitions the process finishes. When it is done, the set of edges it has added forms the minimum spanning tree, or forest if the graph is not connected.
Boruvka Tech stack
- - Google Cloud C2-standard-30 instance
- - Language: C++ implemented from scratch
- - Job Manager: Slurm
- - Parallel Execution Model: Single Program-Multiple Data (SPMD)
- - Type of Application: HPC
- - MPI/OpenMP Hybrids systems
- - Inter-node: MPI (distributed memory model), Data Parallelism
- - Intra-node: OpenMP (shared memory model), Function Parallelism
Unet
UNet is a popular deep neural network that has shown great performance in image segmentation. It is a U-shaped stack of convolutional layers that up-samples a large number of local features and then concatenates with feature maps in the reverse order while downsampling to the image space. Configured with a large number of trainable parameters, it is able to learn from annotated data through epochs of training and accurately determine whether each pixel belongs to a nucleus or not. With data augmentation techniques, this model learns well through only very few epochs.
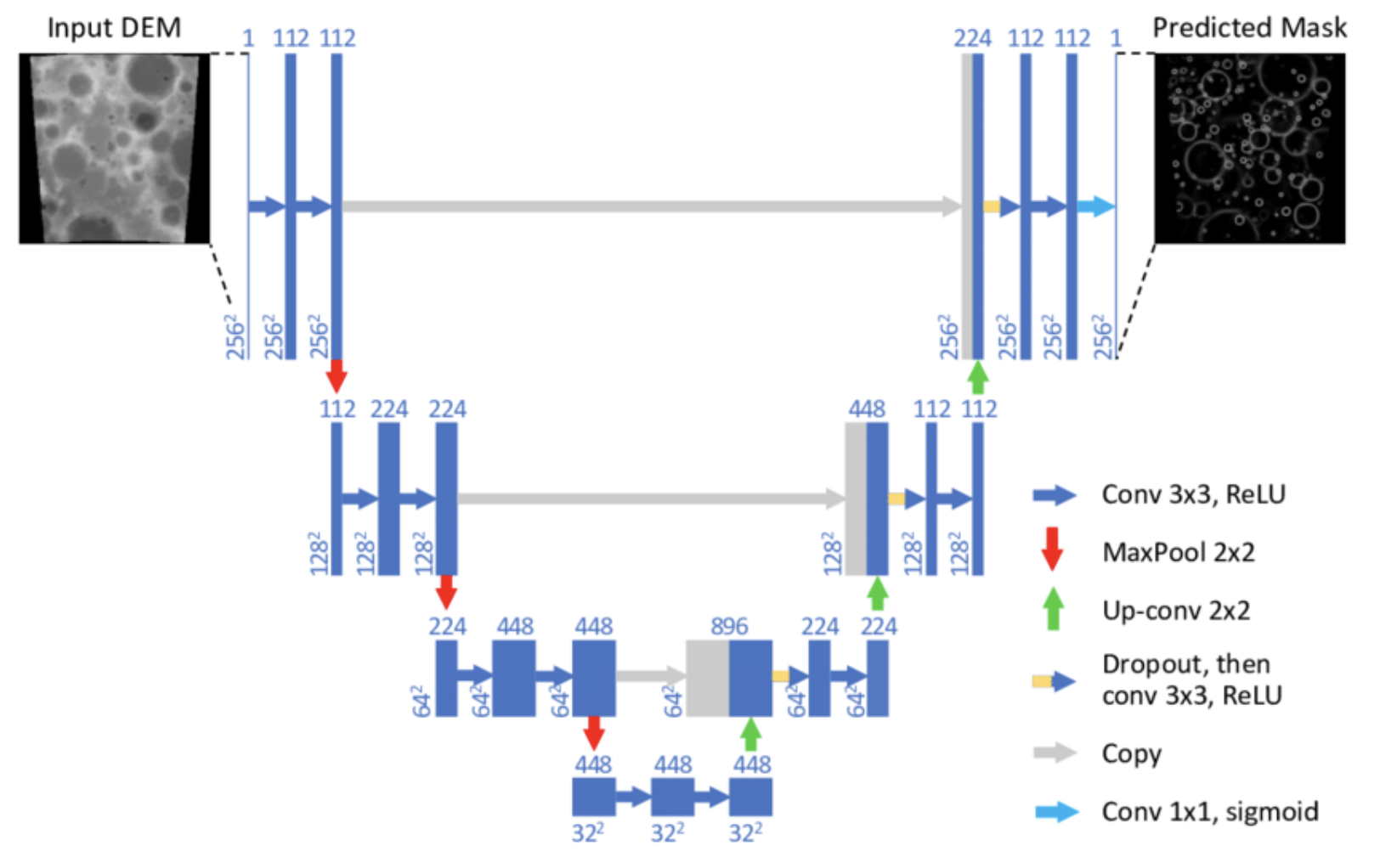
- Highly non-linear function mapping each pixel to 0 or 1
- Huge amount of model parameters
- Total params: 1,941,105
- Trainable params: 1,941,105
- Learns fast with data augmentation
Unet Tech stack
- - Infrastructure:
- Google Cloud Compute Engine and Dataproc cluster
- AWS VM (multiple instance types)
- - GPU accelerated computing
- Single GPU of different types
- Parallel data augmentation and training
- - Storage: Google Cloud Storage bucket
- - Optimization: GPU, Spark
- - Language: Python
- - Software: Keras + Tensorflow
- - Platform: Google Cloud Platform & AWS
- - Levels of Parallelism: Task level
- - Types of Parallelism: Data parallelism automated by GPU
Results
- All
- Otsu
- Boruvka
- Unet
image 1

result 1 - Boruvka
image 2

result 2 - Boruvka
image 3

result 3 - Otsu
image 4

result 4 - Otsu
image 5

result 5 - Unet
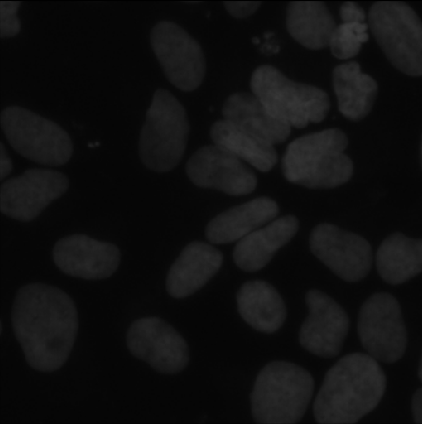
image 6
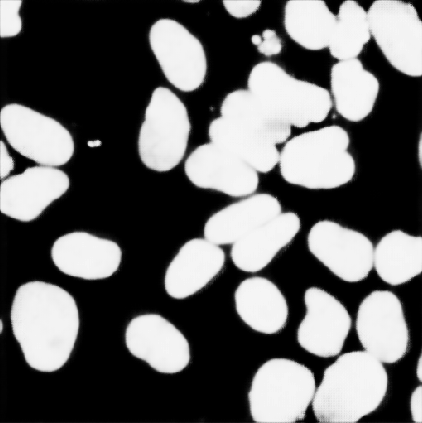
result 6 - Unet
Discussion
1 Accuracy

We compare the output quality of the three methods.
Otsu’s method works for most cases but sometimes cannot distinguish between nuclei and background.
Boruvka’s algorithm generates zig-zag and artistic instance boundaries.
UNet gives the most accurate results, but it takes much longer to train and test compared to the other two methods.
2 Speedup
Features
Google Cloud Platform
We explore creating bucket, lanuching VM instance, installing packages, lanuching cluster on GCP. We think GCP has more detailed documentations compared to AWS and we enjoy $300 free credits:)
Slurm Cluster & MPI
As a cluster workload manager, Slurm provides a framework for starting, executing, and monitoring parallel job on the set of allocated compute nodes. It uses NFS for storage and works well with MPI.
Python & C++
We leverage the strengths of both languages - we use Unet, a well-developed package in Python and also low level language C++ to explore parallelism at more fine-grained level.
TensorBoard
TensorBoard is a profiling tool that monitors tensorflow-based model training. It provides a very detailed breakdown of how much time is spent in each section as well as the model performance across epochs.
Graph & Memory
We are working with 256*256 images, which has 65536 pixels. Running MST algorithm on this connected graph with 65536 vertices needs careful design and consideration of how C++ allocates memory.
Data Dependency & lock-free algorithm
Union and find algorithm itself has great data dependence and thus is hard to parallelize. We read a paper using the "compare and swap" technique and implement it, which mitigates the problem and uses less locks.
Challenges
1. Keras is not thread-safe, so a built-in threading package in python cannot be applied,
2. Keras has a bottleneck in CPU usage due to its image_generator function for data augmentation,
3. Multiple dependencies within the existing python packages.
Future work
Otsu's Method
Split Huge Image
In case we have a huge image, we may parallelize its segmentation by splitting it into smaller chunks for parallel processing.
Boruvka's Method
Feature Aggregation
When looking at some bad outputs produced by Boruvka, we notice that there are some wired vertical lines that should definitely not belong to the place it assigned. We deduct the reason of that happening is that we use fixed weight edges in our program. The weight of edge (between two components) always equals the weight of the edge in the original graph and the color difference between its two endpoints. This infomation is local and error prone! Imagine if we use the color difference between two supernode components to represent edge weight, we get more accurate estimate of color difference between two components and thus will probably get a better and more sensible result.
Unet
Multi-GPU instance
Currently, we are constrained to a single-instance GPU on Google Cloud due to its quota limit. If we are able to obtain more GPU instances, we hope to train the network on multi-GPU with the built-in methods by Keras for distributed model training.
Multi-worker Spark Cluster
We attempted to use Elephas and PySpark for distributed training on a cluster of CPU workers, but perhaps due to the large number of parameters, we kept running into memory error on spark. We may further investigate the reason why it fails.
Data parallelism during inference step
In prediction when there is a large number of test images, we should explore data parallelism such that a pretrained model can be applied to multiple test images concurrently to segment the nuclei and efficiently saves the results.
Acknowledgements
Special thanks to Dr.Sondak and all CS205 TFs! Thank you for the semester!
[1] Sun Chung and A. Condon, "Parallel implementation of Bouvka's minimum spanning tree algorithm," Proceedings of International Conference on Parallel Processing, 1996, pp. 302-308, doi: 10.1109/IPPS.1996.508073.
[2] A. Mariano, A. Proenca and C. Da Silva Sousa, "A Generic and Highly Efficient Parallel Variant of Boruvka's Algorithm," 2015 23rd Euromicro International Conference on Parallel, Distributed, and Network-Based Processing, 2015, pp. 610-617, doi: 10.1109/PDP.2015.72.
[3] Tepper, Mariano & Mejail, Marta & Musé, Pablo & Almansa, Andrés. (2011). "Boruvka Meets Nearest Neighbors". 8259. 10.1007/978-3-642-41827-3_70.
[4] X. Wei, Q. Yang, Y. Gong, N. Ahuja and M. Yang, "Superpixel Hierarchy," in IEEE Transactions on Image Processing, vol. 27, no. 10, pp. 4838-4849, Oct. 2018, doi: 10.1109/TIP.2018.2836300.
[5] Richard J. Anderson and Heather Woll. 1991. "Wait-free parallel algorithms for the union-find problem," in Proceedings of the twenty-third annual ACM symposium on Theory of Computing (STOC '91). Association for Computing Machinery, New York, NY, USA, 370–380. doi:https://doi.org/10.1145/103418.103458
[6] Win KY, Choomchuay S, Hamamoto K, Raveesunthornkiat M. Comparative Study on Automated Cell Nuclei Segmentation Methods for Cytology Pleural Effusion Images. J Healthc Eng. 2018;2018:9240389. Published 2018 Sep 12. doi:10.1155/2018/9240389
[7] Long, F. Microscopy cell nuclei segmentation with enhanced U-Net. BMC Bioinformatics 21, 8 (2020). https://doi.org/10.1186/s12859-019-3332-1
[8] Zhu, Wentao, Zhao, Can, et al. LAMP: Large Deep Nets with Automated Model Parallelism for Image Segmentation. 2020. https://arxiv.org/pdf/2006.12575.pdf
